The last few years have seen an incredible increase in the discovery of genetic variants associated with depression. In this Mythbusters blog, Kirstin [EDIT Lab PhD student] and Joni [SGDP Postdoc] describe this recent progress, and explain why there isn’t just a single gene for depression.
We’ve known for a long time that depression runs in families. We’ve also known for a long time that things that run in families are very likely to be caused, at least in part, by genes. Genes are sections of DNA that encode proteins, which play a major role in human biology. Changes in the DNA can affect the structure of the proteins encoded by genes, or can affect the amount of protein produced. As a consequence, changes in DNA can alter biology, which can alter behaviour. Knowing this led scientists down a long and windy road to understand what role DNA plays in depression and what specific genetic variants are implicated.
Before the landmark Human Genome Project, where scientists sequenced the human genome for the first time (identifying and mapping out all the [nucleotides] and genes on the genome), discovering actual differences in DNA between people with or without depression, or any disorder, was a nearly impossible task. Without a full understanding of how many such differences were present in the genome, scientists had to make educated guess at which genes were involved, and which differences were relevant. The genes that scientists guessed were important are called ‘candidate genes’. Unfortunately, these guesses haven’t turned out to be correct – there is little evidence that any of these candidate genes are involved in depression.
“Scientists turned to the genome-wide association study (or GWAS), a method that allowed them to look at many variants across the genome…“
Instead, scientists turned to the genome-wide association study (or GWAS), a method that allowed them to look at many variants across the genome without prior assumptions about which variants were relevant. Several GWAS were carried out without identifying any significant variants, which suggested any individual variant would only be very weakly associated with having depression. Then, a series of GWAS were published identifying gradually more and more DNA variants associated with depression. First, a study in Chinese women with severe depression identified two such DNA variants where there are differences between cases and controls, and then the direct-to-consumer DNA testing company 23andMe published a GWAS in White European participants that identified 15 such differences. Combining this with data from the Psychiatric Genomics Consortium (an international collaboration of scientists interested in the genetics of psychiatric disorders) yielded 44 regions of the genome associated with depression. Most recently, including data from the UK Biobank (a massive study of the health of UK adults) raised this number to 102 DNA differences associated with depression.
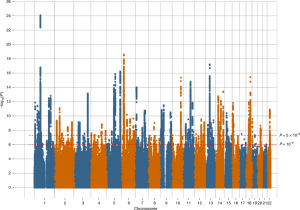
The ‘Manhattan plot’ from the most recent depression GWAS, which shows the significance of the associations between each genetic variant and depression. These plots get their name as the many associated DNA changes across the genome result in the plots resembling the Manhattan skyline.
“Research in depression genetics has demonstrated that there is no “gene for depression””
Research in depression genetics has demonstrated that there is no “gene for depression” – instead there are many DNA changes (probably 1000s) with very small individual associations with depression, which together act to increase an individual’s chances of developing the illness. Genetic discovery in depression is far from over – what variants have been identified only perform a little bit better than chance when separating people who have depression from those who do not, and the ability to predict whether a specific person has depression is still a long way off. Furthermore, examining depression as a single concept might obscure genetic associations with particular symptoms or other aspects of depression. Continuing to identify DNA changes associated with depression or its component parts will allow us to begin to understand the biology of the illness, with the potential to develop new treatments as well as improving those we have. Studies like the GLAD study will be a big part of that, increasing the number of people available to study and so increasing the number of DNA differences identified. Furthermore, depression genetic research to date has largely been restricted to studying White Europeans – we hope GLAD can help to increase the number of BAME participants in genetic studies, and allow our findings to be valuable for everyone.




[…] I use evidence about this to show that depression can be at least partly explained genetically. This piece explains the problem of replicating this evidence. This is highlighted by Thalia Eley in a lecture, an extract of which is also on the webpage. For a more up to date picture of the genetics of depression, please follow this link. […]