The month of October hosts UK Black History Month, World Mental Health Day, and the anniversary of the Human Genome Project. Inspired by all three, Elisavet [EDIT Lab first-year PhD student] and Elena [EDIT Lab 2020-2021 MSc student] discuss the need to diversify genetics research for the continual progress of the mental health field.

Elisavet ( EDIT Lab 1st year PhD student)

Elena (EDIT Lab 2020-2021 MSc Student)
NB: This blog discusses concepts like ancestry, race, and ethnicity. This EDIT Lab blog by Dr Yasmin Ahmadzadeh is a great reference for the distinction between these terms.
Genetics research has developed rapidly over the past 20 years, enabling researchers to identify key genetic variants (i.e. slight differences in the DNA sequence of genes) that are associated with measurable human traits. These traits range from physical characteristics, such as body mass index, to mental health conditions, such as schizophrenia, and their underlying genetic basis is commonly studied using genome-wide association studies (GWASs).
In a GWAS, participants are split into cases (people with the disease, disorder, or trait of interest) and controls (those who do not have the disease, disorder, or trait). Data from these participants are then compared to identify whether any genetic variants are found (statistically) significantly more often in the cases versus the controls. If such genetic variants are detected, this would be considered evidence for their role in the trait of interest.
The past two decades have seen an exponential increase in the number of GWASs conducted (1,2), and as of 2021, more than 5,300 publications have been reported (3). Findings from GWASs can improve our understanding of the genetic basis of various conditions and disorders, and have the potential to be translated into clinical practice. For example, the most recent GWAS of anorexia nervosa provided insight into the genetic overlap between the disorder and other traits with which it often co-occurs, namely: other psychiatric disorders, the tendency for high physical activity, and metabolic and anthropometric traits (4). Taken together, these findings showed that this is a metabolic as well as a psychiatric disorder.
 So, what is the problem?
So, what is the problem?
Large scale GWASs have demonstrated that many common disorders, diseases, and traits are highly complex (or polygenic) and that they are partially influenced by the combined effects of thousands of single genetic variants, each of which may only have a very small influence. Thus, GWAS generally rely on data from tens of thousands or even hundreds of thousands of participants to identify the typically small effects of specific genetic variants. A key source of genomic data comes from biobanks (5), and these large-scale databases contain genetic and health data from millions of people. Over the past 30 years, more than 100 biobanks have been established worldwide, with the UK Biobank being one of the largest, storing data for half a million people.
The open-access nature of biobanks has led to a tidal wave of studies using their data (6), and enabled the discovery of many key genetic variants involved in physical and mental health conditions. However, this data is not representative of global ethnicities (e.g. 95% of UK Biobank is currently composed of white participants (7)).
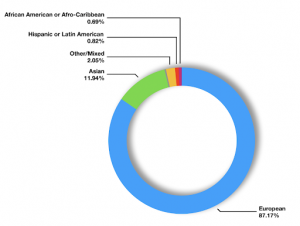
Currently, more than 70% of participants in GWASs are both of European descent and from just three countries: the US, the UK, or Iceland, with the UK being the largest contributor (8). This becomes problematic as the majority of the world’s population (76%) reside in Asia or Africa, but data from non-white participants (i.e. the 5% of the UK Biobank) are used at a minimum rate in GWAS or genetics research, in comparison to white samples. For example, in 2020, only 10% and less than 1% of GWAS participants were of Asian and admixed African descent, respectively (see pie chart below) (9).
Figure 1. The percentage of individuals from each ancestral population included in genome- wide association studies, as of the 27th of October and as reported by the real-time Diversity Monitor (12).
Why is this a problem?
The primarily white samples, consisting largely of sub-populations of European descent from the UK and the US, are not representative of the global population. In fact, studies highlight that traits are differently associated with genetic variants in different ancestry groups (10,11). Therefore, the findings from GWAS and, in turn, any benefits generated from them in terms of treatment, detection, or prevention of disease, currently apply only to individuals of specific ancestry groups. As we aim to better understand, identify, prevent, and treat complex conditions, this leaves us with a story half-told.
At this point, it is important to emphasise that the identified genetic differences across ancestry groups do not represent different racial categories (refer to (8) for a detailed discussion).
There are multiple explanations for the differences in genetic variation between ancestry groups (13), but expanding upon them is beyond the scope of this blog. For those interested, the linked papers on linkage disequilibrium and population stratification explain how they may influence GWAS findings.
How can diversity in GWAS improve research and clinical outcomes?
Including ancestrally diverse samples in GWASs would enable more accurate and reliable implementation and expansion of the current findings to identify, research, and ultimately treat common and complex traits, such as mental health conditions, for a wider range of people.
For example, a GWAS on schizophrenia that included genomic data from individuals of admixed African, Latinx, and European ancestry improved our confidence in the previously and newly identified genetic variants associated with the risk for the disorder, not only for the individuals of the understudied populations but also for those of European descent (14). Notably, the associations with African-derived genetic variants were generalisable to both European and Latinx ancestries, highlighting the significant contribution of diverse GWAS research not only within but also beyond ancestral groups. This valuable information from each population can help geneticists to disentangle complex signals and pinpoint the genes that functionally impact risks for disorders and diseases.
In clinical practice, diverse ancestry GWAS will also allow for the discovery of biological effects that are too rare to be discovered in European samples, but important for drug development. This is meaningful as most drugs have been based on discoveries in European populations, but work equally well for most populations. Thus, studying many global populations is likely to reveal new mechanisms and provide evidence for more new treatments more quickly than if we concentrate on just one population.
Conducting this research may also elucidate the role of ancestry in the risk for disease. For example, African ethnicity is considered a risk factor for psychiatric illnesses in the UK, especially for schizophrenia (15). More genetics research into this (from UK populations of African ancestry) will be vital in treating and understanding the aetiology of such conditions (e.g. whether there are particular genetic risks that are unique to or stronger in people of African ancestry, or whether this disparity reflects socioeconomic differences or medical racism).
Diversifying genetics research can thus address and transcend health inequalities on various levels by ensuring the medical relevance of GWAS findings for understudied populations. Thus, moving forward, this pursuit could improve the translation of genomic research into clinical practice, precision medicine, and public health policy that equally benefit all, not just a select few.
In Part 2, we discuss the way that changes toward diverse ancestry research can be enacted at the level of funding bodies, academic and research institutions, and individuals.
References
- MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2016/11/29 ed. 2017 Jan 4;45(D1):D896–901.
- Loos RJF. 15 years of genome-wide association studies and no signs of slowing down. Nature Communications. 2020 Nov 19;11(1):5900.
- National Human Genome Research Institute. GWAS Catalog [Internet]. The NHGRI-EBI Catalog of published genome-wide association studies. 2021 [cited 2021 Oct 18]. Available from: https://www.ebi.ac.uk/gwas/labs/
- Watson HJ, Yilmaz Z, Thornton LM, Hübel C, Coleman JRI, Gaspar HA, et al. Genome-wide association study identifies eight risk loci and implicates metabo-psychiatric origins for anorexia nervosa. Nat Genet. 2019 Aug;51(8):1207–14.
- Illumina, Nature Research Media Custom Media. Population matters: Biobanks accelerate geno–pheno discoveries [Internet]. Nature Portfolio. 2020. Available from: https://www.nature.com/articles/d42473-020-00238-1
- Claussnitzer M, Cho JH, Collins R, Cox NJ, Dermitzakis ET, Hurles ME, et al. A brief history of human disease genetics. Nature. 2020 Jan;577(7789):179–89.
- Davis K, Coleman J, Adams M, Allen N, Breen G, Cullen B, et al. Mental health in UK Biobank: development, implementation and results from an online questionnaire completed by 157 366 participants – CORRIGENDUM. BJPsych Open. 2018;4(5):352–3.
- Mills MC, Rahal C. A scientometric review of genome-wide association studies. Communications Biology. 2019 Jan 7;2(1):9.
- Home – GWAS Diversity Monitor [Internet]. [cited 2021 Oct 25]. Available from: https://gwasdiversitymonitor.com/
- Carlson CS, Matise TC, North KE, Haiman CA, Fesinmeyer MD, Buyske S, et al. Generalization and Dilution of Association Results from European GWAS in Populations of Non-European Ancestry: The PAGE Study. PLOS Biology. 2013 Sep 17;11(9):e1001661.
- Dunn EC, Sofer T, Wang M-J, Soare TW, Gallo LC, Gogarten SM, et al. Genome-wide association study of depressive symptoms in the Hispanic Community Health Study/Study of Latinos. Journal of Psychiatric Research. 2018 Apr 1;99:167–76.
- Mills MC, Rahal C. The GWAS Diversity Monitor tracks diversity by disease in real time. Nature Genetics. 2020;52:242–3.
- Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019 Apr;51(4):584–91.
- Bigdeli TB, Genovese G, Georgakopoulos P, Meyers JL, Peterson RE, Iyegbe CO, et al. Contributions of common genetic variants to risk of schizophrenia among individuals of African and Latino ancestry. Molecular Psychiatry. 2020 Oct 1;25(10):2455–67.
- Fearon P, Kirkbride JB, Morgan C, Dazzan P, Morgan K, Lloyd T, et al. Incidence of schizophrenia and other psychoses in ethnic minority groups: results from the MRC AESOP Study. Psychological Medicine. 2006 Nov;36(11):1541–50.

[…] Part 1 of this blog series, Elisavet [EDIT Lab first-year PhD student] and Elena [EDIT Lab 2020-2021 MSc […]