This week in our A-Z post for ‘K’, Tom McAdams [EDIT Lab Associate Director] outlines Kinship, a core concept in the field of behavioural genetics.
‘Kinship’ can have different meanings to scientists from different fields, but to those interested in genetics, it typically refers to the degree of genetic relatedness between individuals. This is often expressed as a relationship coefficient giving the proportion of genetic variance shared between two individuals. By “proportion of genetic variance”, we mean the proportion of the human genome that can differ between people. For example, monozygotic (identical) twins share 100% of their DNA, expressed as 1.00. Dizygotic (fraternal) twins, as well as siblings, share on average 50% of their genetic variance, so they are said to have a genetic relatedness coefficient of .50. For half-siblings it is .25, for cousins it is .125, and so on. That is, for each degree of separation, the relatedness coefficient halves.
“[Kinship] is often expressed as a relationship coefficient giving the proportion of genetic variance shared between two individuals”
Why is this?
Parents pass a random half of their DNA on to each of their children. This is why parents share 50% of their genetic variance with their children. If a mother has two children, she will pass on a random half of her DNA to each child. The two children will share (on average) 25% of their maternal DNA (i.e. they will inherit a different random half of their mother’s DNA). If the two children have different (and unrelated) fathers, then this shared maternal DNA will be the extent of their shared genetic variance – they will be half-siblings, with a relatedness coefficient of .25. If the two children share a father as well as a mother, then they will also share 25% of their paternal DNA. Combined with their maternal DNA, this means that they will share 50% of their genetic variance, giving a relatedness coefficient of .50.
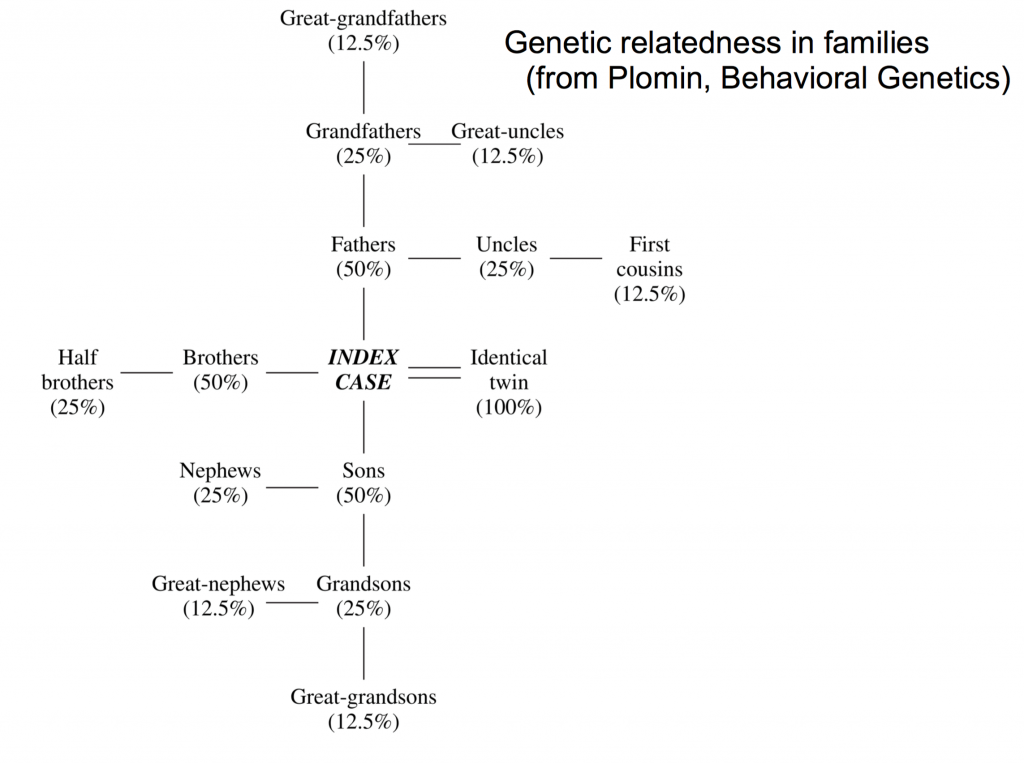
Parents, siblings and offspring are considered first-degree relatives, and all share 50% of their genetic variance. Second degree relatives include half-siblings, grandchildren, grandparents, aunts/uncles, and nieces/nephews, and share 25% of their genetic variance. Third-degree relatives include great-grandparents, great-uncles/aunts (the siblings of grandparents), cousins, and great-grandchildren, and share 12.5 % of their genetic variance. The diagram below illustrates this point.
In reality, there will of course be some variation around these relatedness coefficients. For example, in a sample of sibling pairs, by chance some pairs will share slightly more than 50% of their genetic variance, and some will share slightly less, but the mean average will be 50%. This average could however be pushed upwards in certain circumstances. For example, if there is a lot of in-breeding within a population, then parents may be related to one another, meaning that siblings would tend to share more than 50% of their genetic variance.
“As well as sharing genes, relatives often share their environment.”
What about environmental relatedness?
As well as sharing genes, relatives often share their environment. Siblings growing up in the same house share a nuclear family environment. Parents and children share a nuclear family environment as well. Cousins do not share a nuclear family environment, but they may share an extended family environment.
“we can use what we know about their[twins] genetic and environmental relatedness to calculate heritability estimates”
Why does any of this matter?
Knowledge regarding genetic and environmental relatedness can be used to establish how important genetic and environmental influences are in explaining population variance on a trait. For example, if a sample of identical twins and a sample of non-identical twins are assessed in terms of their IQ, and it is found that the identical twins are more similar to one another than are the non-identical twins, then we can use what we know about their genetic and environmental relatedness to calculate e.g. heritability estimates (as covered in more detail here). This is the basis of the twin method for calculating heritability, but the same logic can also be applied to comparisons between siblings and cousins, half-siblings and half-cousins.
This is what makes the concept of kinship so important to behavioural geneticists. Comparisons between samples of differentially related individuals provide us with a useful tool with which to evaluate the aetiology of complex traits.