In the last few decades, non-coding DNA has been a hot topic of discussion and debate within the scientific community. In this blog, we decode the mysteries of non-coding DNA, unveiling their genetic significance beyond the “junk” label.
Figure from: https://geneticliteracyproject.org/2021/09/17/junk-dna-the-98-of-the-human-genome-that-does-not-encode-proteins-is-often-called-useless-but-the-reality-is-more-complicated/
The discovery of the structure of DNA by James Watson and Francis Crick in 1953 was a milestone in the field of biology, marking a turning point in the history of genetics (Watson & Crick, 1953). Subsequent advances in molecular biology revealed that out of the 3 billion base pairs of human DNA, only around 2% codes for proteins; many scientists argued that the other 98% seemed like pointless bloat of genetic material and genomic dead-ends referred to as non-coding DNA, or junk DNA – a term you’ve probably come across (Ohno, 1972).
First coined in the 1960s and later popularised by geneticist and evolutionary biologist Susumu Ohno in 1972, the term junk DNA has stuck for many years. Ohno first described that large genomes would inevitably harbour sequences accumulated over several years that did not encode any proteins, stating: “The earth is strewn with fossil remains of extinct species; is it a wonder that our genome too is filled with the remains of extinct genes?” (Ohno, 1972). Soon after his statements, scientists discovered evidence of just how prevalent and diverse non-coding DNA is in our genome.
The concept of non-coding DNA continued to be a shrouded mystery, with questions lingering over its true value: is it merely just junk, or something more meaningful? The debate could be summarised as “Only regions of DNA conserved across species are important” versus “If non-coding DNA is “junk”, it would have been removed from the genome during evolution.” In the last decade new research findings – thanks to technological advancements – have compelled both sides of the argument to reconsider their views on non-coding DNA.
One of the most significant studies to challenge the notion of non-coding DNA was the ENCODE (ENCyclopedia Of DNA Elements) project (The ENCODE Project Consortium, 2012). First launched in 2003, this project aimed to investigate the role of non-coding regions in the genome. In 2012, the project released a series of papers discovering that around 75% of the non-coding DNA in the human genome underwent transcription (i.e., the process of making copies of DNA parts that contain certain instructions so that our cells can read them and use them to carry out their functions). Moreover, they found that 50% of the genome was accessible to proteins involved in genetic regulation, such as transcription factors. Transcription factors are proteins that help control which parts of our DNA are used to make different types of cells and tissues in our bodies, like light switches that can turn genes on or off depending on what cells need. ENCODE researchers also estimated that at least 80% of the genome had biochemical activity, suggesting that much of the non-protein-coding parts may have regulatory roles (The ENCODE Project Consortium, 2012). However, these findings were challenged by other scientists who claim that just because these genomic segments have access to transcription factors does not mean they have any biochemical function, and transcribing these regions is not advantageous in terms of evolution. Nonetheless, since 2012, a series of studies have gradually illuminated the function role of non-coding DNA.
Below are three examples of studies of non-coding DNA relating to mental health disorders, including non-coding RNAs, telomeres and alternative splicing:
- Non-coding RNAs: Cells use non-coding DNA to produce various types of RNA molecules following transcription. RNA takes the instructions from our DNA and brings them to the parts of the cell where they can be used to make proteins. Non-coding RNAs (ncRNAs) are RNAs that are not translated to produce proteins but still have many regulatory roles. ncRNAs expressed in the brain have been implicated in the onset of major depressive disorder, schizophrenia, and bipolar disorder (Yoshino & Dwivedi, 2020). For example, studies have found a difference in the number of ncRNAs expressed in depressed patients in comparison to controls, with evidence suggesting that antidepressants can alter the aberrant expression of ncRNAs in depressed patients (Lin & Turecki, 2017).
- Telomeres: A significant and variable portion of non-coding DNA in genomes consists of highly repeated sequences. Telomeres, for instance, are mostly made up of these repeated sequences and are thought to help maintain chromosome integrity (shortening telomeres have been linked to ageing). Studies have found that older adults with late-life depression have shorter telomeres than controls, especially those with a more severe depressive episode, suggesting that shorter telomeres can be a marker of the severity of depressive episodes (Mendes‐Silva et al., 2021).
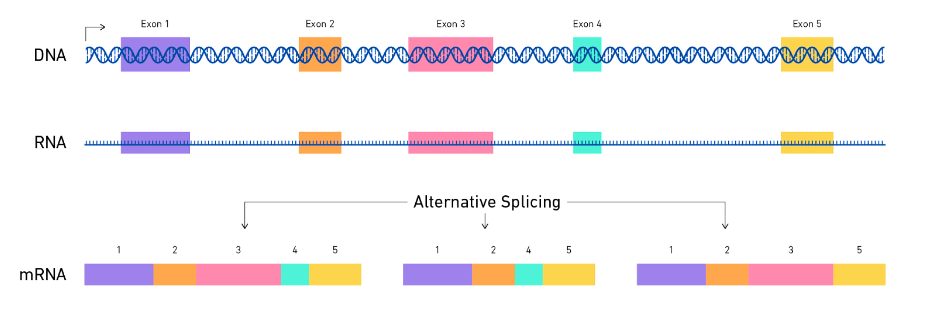
- Alternative splicing: Alternative splicing is a process by which the same gene can produce different mRNA transcripts by selectively including or excluding certain regions of the DNA sequence, including non-coding regions. To increase expression of a gene that regulates serotonin (HTR1A) splicing removes a specific site from the RNA molecule that regulates its stability, resulting in more stable RNA. However, this splicing process is different in various regions of the brain, and studies have found it to be less efficient in people with major depression. Moreover, certain factors that control this splicing process for HTR1A RNA were also found to be reduced in people with depression. This specific non-coding region may contribute to attracting a destabilising complex that eventually results in lower levels of the serotonin receptor in the cortex (Le François et al., 2018).
Figure from: https://www.technologynetworks.com/genomics/articles/alternative-splicing-importance-and-definition-351813
Non-coding DNA has also become increasingly important in genome-wide association studies (GWAS), which aim to identify genetic variants associated with diseases or traits. While GWAS studies initially focused on coding regions of the genome, researchers have now discovered that many significant single nucleotide polymorphisms (SNPs) identified in GWAS studies actually fall within non-coding DNA, most often in introns of genes, highlighting the importance of studying these once-ignored regions of the genome in the context of disease genetics (Levings et al., 2020; Ormel et al., 2019).
Foods considered to have very little nutritional value are called junk food, or piles of email advertisements are referred to as junk mail. The term “junk DNA” implies it is just a non-coding filler of useless sequences that can inevitably be disregarded. However, growing bodies of evidence suggest that genetic variation in them has health implications. Nowadays, researchers are less likely to describe any non-coding sequences as junk because there are multiple other and more accurate ways of labelling them. The discussion over non-coding DNA’s function is not over, and it will be long before we understand our whole genome. For many researchers, the field’s best way ahead is keeping an open mind when evaluating the functional consequences of non-coding DNA and RNA, and not to make assumptions about their biological importance.
References:
Le François, B., Zhang, L., Mahajan, G. J., Stockmeier, C. A., Friedman, E., & Albert, P. R. (2018). A Novel Alternative Splicing Mechanism That Enhances Human 5-HT1A Receptor RNA Stability Is Altered in Major Depression. The Journal of Neuroscience, 38(38), 8200–8210. https://doi.org/10.1523/JNEUROSCI.0902-18.2018
Levings, D., Shaw, K. E., & Lacher, S. E. (2020). Genomic resources for dissecting the role of non-protein coding variation in gene-environment interactions. Toxicology, 441, 152505. https://doi.org/10.1016/j.tox.2020.152505
Lin, R., & Turecki, G. (2017). Noncoding RNAs in Depression. In R. Delgado-Morales (Ed.), Neuroepigenomics in Aging and Disease (Vol. 978, pp. 197–210). Springer International Publishing. https://doi.org/10.1007/978-3-319-53889-1_11
Mendes‐Silva, A. P., Vieira, E. L. M., Xavier, G., Barroso, L. S. S., Bertola, L., Martins, E. A. R., Brietzke, E. M., Belangero, S. I. N., & Diniz, B. S. (2021). Telomere shortening in late‐life depression: A potential marker of depression severity. Brain and Behavior, 11(8). https://doi.org/10.1002/brb3.2255
Ohno, S. (1972). So much ‘junk’ DNA in our genome. Brookhaven Symposia in Biology, 23, 366–370.
Ormel, J., Hartman, C. A., & Snieder, H. (2019). The genetics of depression: Successful genome-wide association studies introduce new challenges. Translational Psychiatry, 9(1), 114. https://doi.org/10.1038/s41398-019-0450-5
The ENCODE Project Consortium. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature, 489(7414), 57–74. https://doi.org/10.1038/nature11247
Watson, J. D., & Crick, F. H. C. (1953). Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid. Nature, 171(4356), 737–738. https://doi.org/10.1038/171737a0
Yoshino, Y., & Dwivedi, Y. (2020). Non-Coding RNAs in Psychiatric Disorders and Suicidal Behavior. Frontiers in Psychiatry, 11, 543893. https://doi.org/10.3389/fpsyt.2020.543893